I'm Raunak, a master's student at the University of Illinois, Urbana-Champaign. This summer, I had the opportunity to intern as a Software Engineer at LanceDB, an early-stage startup based in San Francisco.
LanceDB is a database company, specializing in the storage and retrieval of multimodal data at scale. I worked on the open-source file format (Lance). The main codebase is in Rust, built on top of Apache Arrow, with a user-facing API in Python. Working at Lance has been a great learning experience that has broadened my perspective and increased my confidence as an engineer.
I worked on several features intended to improve the extent of compression and reduce the read time of data, for both full scans and random access. I got extensive hands-on experience working within the Apache Arrow ecosystem, doing asynchronous programming in Rust, and was able to delve deep into the internals of a real database system, allowing me to understand things at a fundamental level.
A significant portion of my work involved implementing compressive encodings that intelligently processed data at a bit level. For instance, previously strings were encoded using a logical encoding - more specifically, as a List of bytes. My initial goal was to add dictionary encoding to improve string compression. This required first re-implementing the basic binary (string) encoding as a physical encoder from scratch (PR). In the new encoder, I encoded the bytes and offsets of the string array separately. For example:
StringArray: ["abc", "d", "efg"]
Physical Binary Encoding:
Bytes: ['a', 'b', 'c', 'd', 'e', 'f', 'g']
Offsets: [0, 3, 4, 7]Once this was done, I added dictionary encoding (PR). Since a large dictionary size can be suboptimal, this encoding is only used for columns with low cardinality.
StringArray: ["abc", "d", "abc", "d", "efg"]
Dictionary Encoding:
BinaryEncoding(["abc", "d", "efg"])
Indices: [0, 1, 0, 1, 2]When applied, these encodings significantly improved Lance’s performance on string data. On the TPCH dataset, the size of the relevant string columns was reduced by 10–20x, and the read time was lowered by 2–3x. Here is a comparison of how the read times reduced, along with the performance of parquet on the same data:
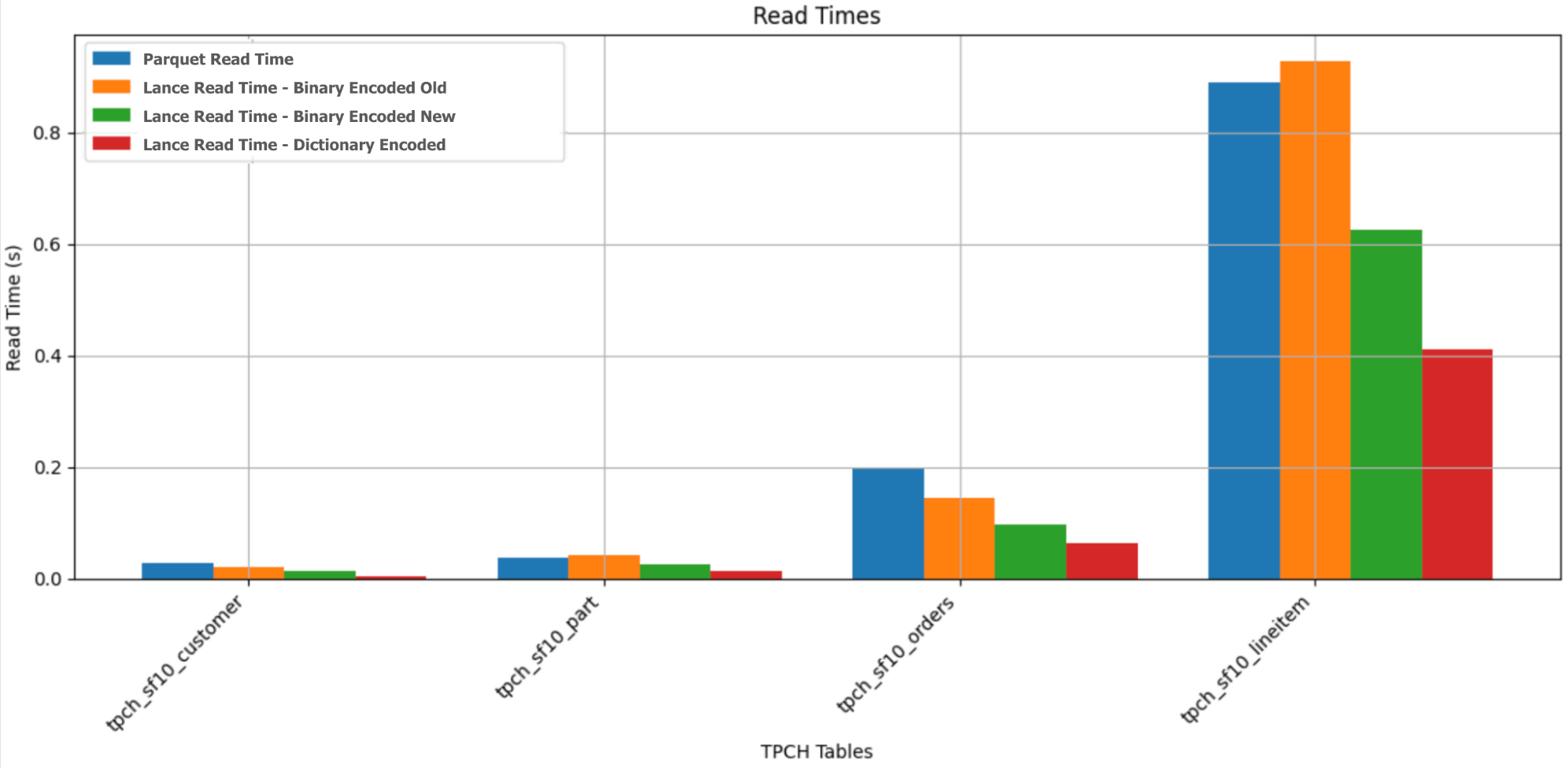
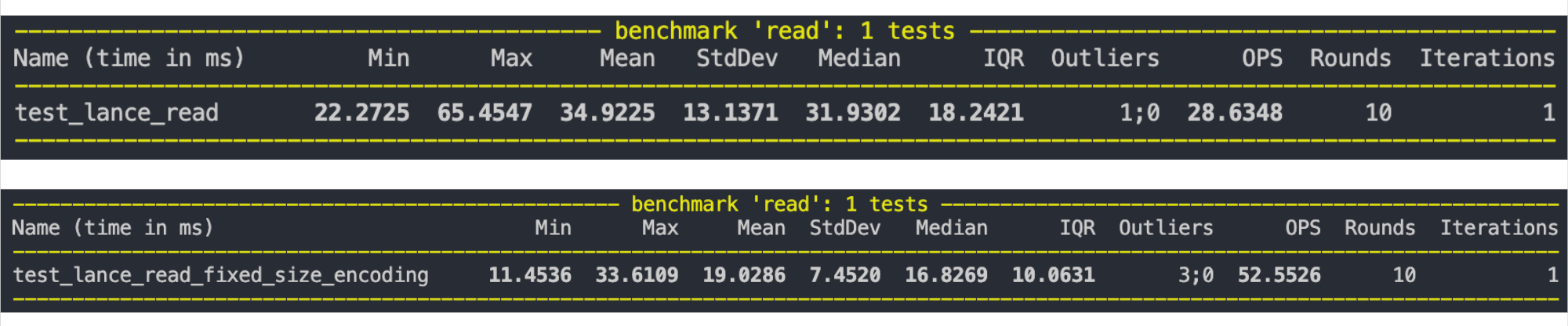
Later, I also added a fixed-size binary encoding (PR). If the strings are of fixed size, we don’t need to store all the offsets. Storing the byte width is enough. This can help save an I/O call (IOP) and result in extra compression when decoding.
StringArray: ["abc", "def", "ghi"]
FixedSizeBinary Encoding:
Bytes: ['a','b','c','d','e','f','g','h','i']
Byte width: 3On running a benchmark on 10M random strings with a fixed size of 8, the file sizes were reduced by around 2x. The read time reduced by almost 2x as well:
Since most workflows are built on top of the file format, optimizing read and write time for maximum performance is critical. Getting such results was also a consequence of benchmarking low-level Rust code, identifying bottlenecks in performance, and optimizing code by using zero-copy operations, all skills that I honed during the internship.
I also worked on a somewhat nontraditional encoding for structs. Previously, retrieving data from a specific sub-field of a struct resulted in a separate IOP. Thus, retrieving data from multiple struct fields incurred multiple IOPs, which was bad for random access. The packed struct encoding (PR) changes this so that the data for all struct fields in a single row can be packed together.
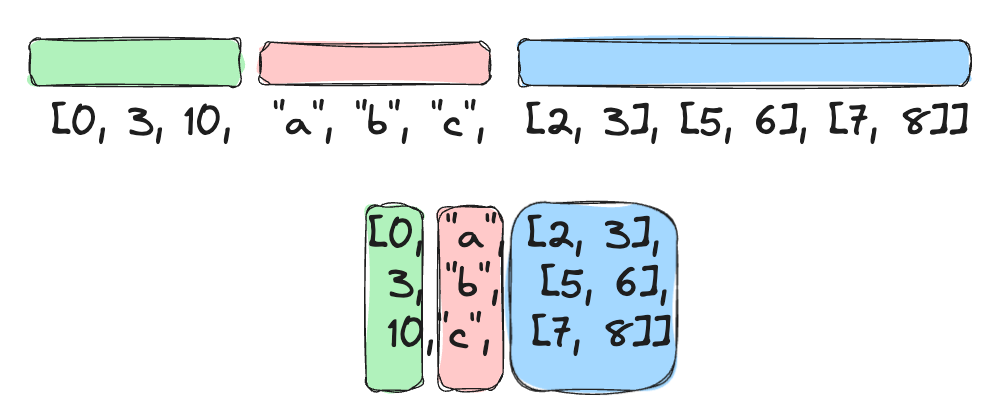
Thus, retrieving even multiple struct fields only consumes a single IOP. This makes random access much faster. On benchmarking a random struct column, we were able to achieve over 3x speedup in average read time:
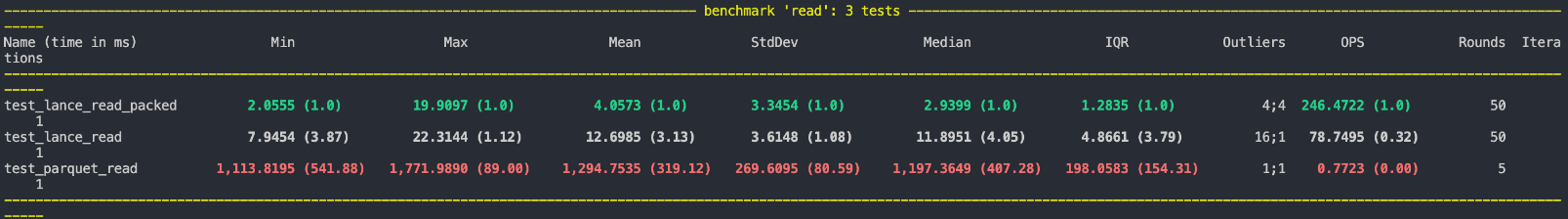
Using this encoding has a trade-off since retrieving an individual struct field will now be slower than before. It should only be applied when the user wants to retrieve multiple struct fields together. This can be indicated by adding a packed: true flag to the struct column metadata.
Implementing such encodings was tricky at first - I had to wrap my head around how bits across multiple nullable arrays were arranged and figure out which bits to decode depending on which rows were queried. Dealing with all the buffer and byte slicing operations while debugging Rust lifetime issues did not make it any easier :) However, my manager was very helpful throughout this process and answered any questions I had. Over time, things got easier. I was able to make faster progress and see improvements in the quality of my work.
Apart from encodings, it was also fun to do some work on the indexing side of things. For example, I added a scalar bitmap index to help search through a column quickly (PR). This was useful for users interested in fast pre-filtering of data. For example, when running a query like SELECT COUNT(*) WHERE age > 30, using a bitmap index on the age column can help speed things up. I also added tooling to help split, shuffle, and load IVF-PQ vector indices for very large-scale datasets (PR1, PR2, PR3). These can be useful for vector search, which is an important use case for enterprises, especially those dealing with storage and retrieval (e.g. RAG) in the GenAI space.
One of the most rewarding aspects of this internship was the opportunity to contribute to an open-source project within a corporate setting. Since the code is public, I can see how the impact of my work (and the project as a whole) evolves over time as the community grows and more developers contribute.
For instance, our dictionary encoding implementation sped up even further after a community contribution, when a user used the hyperloglog approximation for cardinality estimation (this is used in a check to decide whether to use dictionary encoding or not).
Another example was when we found and fixed a bug reported by a user - previously, when randomly accessing multiple rows, each request for a row was scheduled separately. In this PR, I coalesced multiple read requests based on how close the requested rows were, reducing the number of decode tasks being created. When fixed, this helped recover a 10x speed gap on random access.
An open-source format also makes outside collaborations possible. Earlier in June, I presented a research paper at the SIGMOD 2024 conference, where I got a chance to speak with the authors of FastLanes. This eventually led to discussions on integrating FastLanes with Lance. This is still a work in progress, but it can lead to the addition of optimized versions of encodings like delta, frame-of-reference, bit-packing, and more - all of which can significantly improve the performance of the Lance file format.
There are a lot of exciting directions that the Lance file format can take in the future. There are more extensive random access benchmarks that can be added. There are many more encodings we can add to improve performance. Some promising options include a delta encoding, a run-length encoding, or a packed struct encoding for variable-length fields. Experimenting with ALP (Adaptive Lossless Floating-Point Compression) also offers potential benefits.
Another interesting direction is allowing the user to encode a column using multiple encodings to support different use cases. For example, automatically using a packed struct encoding for random access, and an unpacked encoding for full table scans. This would result in faster performance at the cost of some extra storage.
Ultimately, interning at an early-stage startup is a fun experience in itself. The team is extremely efficient, and the culture encourages you to be independent, move fast, and get things done. It was a great opportunity to interact with leaders in the industry and get a glimpse of what really goes into building a tech company.
It's been an exciting 3 months! I've learned a lot, and everyone has been very supportive. Exploring SF and the Bay Area has also been fun. Next, I will be returning to UIUC to complete the final year of my master's degree. I'm looking forward to the adventures that lie ahead!


