In Lance v0.16.1 we introduced several new features, implemented by a combination of LanceDB engineers and community contributors. Lance is an OSS project that is open-to-contribution, so we are very pleased to have major features brought by community members.
In this blog post, we’ll highlight some of the most important new features, which include:
- Version tagging
- Update subschemas in
merge_insert - New file format versioning API
- Distributed ANN index creation API
Version tags
Lance uses multi-version concurrency control, which means each change you make to the dataset creates a new version. Many of these versions are temporary, and later cleaned up. But some you might want to keep around for a while and give a name. With the new Tags feature, you can now do this.
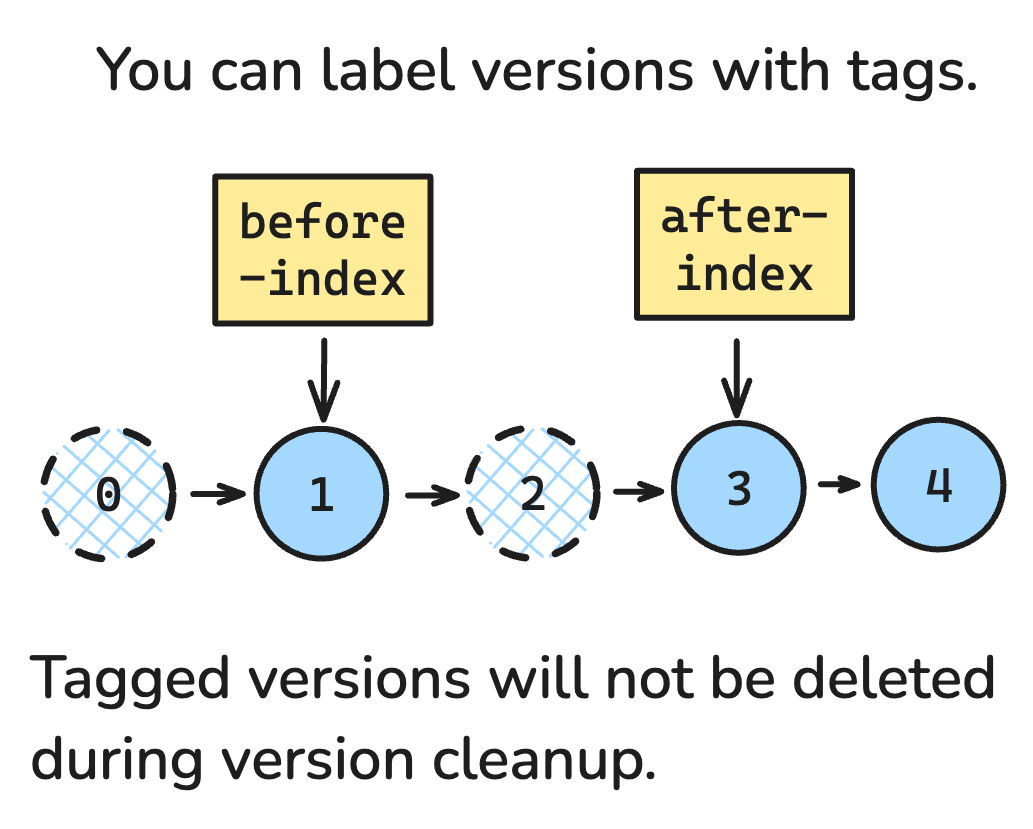
Any version can be given one or more tags. Once tagged, a version of the dataset can’t be deleted, unless you delete the tag. When you load a dataset, you can pass the tag name to load that version.
For example, we can create three versions of a dataset by applying three write operations:
import pyarrow as pa
import lance
assert lance.__version__ == "0.16.1", "wrong version: %s" % lance.__version__
data = pa.table({"x": [1, 2, 3]})
ds = lance.write_dataset(data, "dataset")
ds = lance.write_dataset(data, "dataset", mode="append")
ds = lance.write_dataset(data, "dataset", mode="overwrite")
ds.version
3
We can label the first two versions with a tag. The first one we label as original and the second we label as duplicated. Now we can pass the duplicated tag when opening a dataset to see the version where we duplicated some of the data:
ds.tags.create("original", 1)
ds.tags.create("duplicated", 2)
lance.dataset("dataset", version="duplicated").to_table()
pyarrow.Table
x: int64
----
x: [[1,2,3],[1,2,3]]
We can also pass the original tag to the checkout_version method:
ds.checkout_version("original").to_table()
pyarrow.Table
x: int64
----
x: [[1,2,3]]
We hope these tags open up new workflows for working with datasets and managing versions. We currently support creating and deleting tags. But we also have an open good-first-issue to add support for updating tags too, which could unlock other use cases. For example, imagine maintaining a production tag that is live to users, allowing you to perform operations on your dataset and validate the correctness and performance first before updating the tag to point to a new version.
Community contributor dsgibbons (GitHub username) implemented this new tags feature. Big thanks for their very high quality work. (PR)
Update subcolumns with merge_insert
Merge insert lets you merge new data into a table, inserting new rows and updating existing row. However, right now, you need to supply all the columns in your new data. Sometimes, you want to update a subset of columns.
One use case is where you have a table of documents, and you have a metrics column like “popularity” or “view” count. You would like to update those metrics, but don’t want to have to re-insert the documents or vectors. In Lance v0.16.1, you can do just this! Pass only the match on column and those you wish to update, and it will update only the subset of columns.
documents = pa.Table.from_pylist([
dict(id=1, text="hello", popularity=100),
dict(id=2, text="beautiful", popularity=5_000),
dict(id=3, text="world", popularity=1_000),
])
ds = lance.write_dataset(documents, "documents", mode="overwrite")
ds.to_table()
pyarrow.Table
id: int64
text: string
popularity: int64
----
id: [[1,2,3]]
text: [["hello","beautiful","world"]]
popularity: [[100,5000,1000]]
Updating the popularity column:
new_metrics = pa.Table.from_pylist([
dict(id=1, popularity=200),
dict(id=2, popularity=10_000),
dict(id=3, popularity=2_000),
])
ds.merge_insert(on="id").when_matched_update_all().execute(new_metrics)
ds.to_table()
pyarrow.Table
id: int64
text: string
popularity: int64
----
id: [[1,2,3]]
text: [["hello","world","!"]]
popularity: [[200,10000,2000]]
This is an optimized code path that will only rewrite the relevant columns, even if it affects all rows. This can be a huge savings in AI data, where unstructured data like text or vector embeddings would be expensive to rewrite. For example, we can show a 5,000-fold speedup in rewriting a metric column in a vector dataset with 1 million 1536-dimensional vectors:
# 1 million 1536 dimensional 32-bit vectors
import pyarrow.compute as pc
dim = 1536
def next_batch(batch_size, offset):
global i
values = pc.random(dim * batch_size).cast('float32')
return pa.table({
'id': pa.array([offset + j for j in range(batch_size)]),
'vector': pa.FixedSizeListArray.from_arrays(values, dim),
'metric': pc.random(batch_size),
}).to_batches()[0]
def batch_iter(num_rows):
i = 0
while i < num_rows:
batch_size = min(10_000, num_rows - i)
yield next_batch(batch_size, i)
i += batch_size
schema = next_batch(1, 0).schema
ds = lance.write_dataset(batch_iter(1_000_000), "vectors", schema=schema, mode="overwrite", data_storage_version="2.0")
Previously, you would have to rewrite all columns, which takes over 50s:
%timeit ds.merge_insert(on="id").when_matched_update_all().execute(pa.RecordBatchReader.from_batches(schema, batch_iter(1_000_000)))
52.9 s ± 4.26 s per loop (mean ± std. dev. of 7 runs, 1 loop each)
Now you can just update the metric column in 10ms, which is about 5,000x times faster:
def narrow_iter(num_rows):
for batch in batch_iter(num_rows):
yield batch.select(["id", "metric"])
update_schema = schema.remove(schema.get_field_index("vector"))
reader = pa.RecordBatchReader.from_batches(update_schema, narrow_iter(1_000_000))
%timeit ds.merge_insert(on="id").when_matched_update_all().execute(reader)
9.73 ms ± 776 µs per loop (mean ± std. dev. of 7 runs, 1 loop each)
We hope this feature unlocks new use cases for storing metrics along side vectors in Lance and LanceDB.
This was implemented by Will Jones (me!). (PR)
V2 format versioning API
As we stabilize the current V2 file format, we are establishing a versioning scheme for our data files. Previously, we provided a use_legacy_format flag to select between V1 and V2 files. But in order to allow continuously rolling out new features in the file format, we'll be providing finer grain version selection. We are close to stabilizing a 2.0 format, and have moved current unstable features into the 2.1 format. The current format versions are:
- 0.1 (legacy): The original Lance file format
- 2.0 (stable): The currently stable V2 format
- 2.1 (next): V2 format with FSST and bitpacking
As we roll out new features in the format, we’ll be adding new versions. An up-to-date version of this version list will be maintained in our docs at: https://lancedb.github.io/lance/format.html#file-version
When creating new dataset, you can select any of these versions. The legacy / 0.1 version remains the default for now, until we announce the full stability of the 2.0 format. You can either request the file format version with a label ("stable", "legacy", or "next"):
ds_legacy = lance.write_dataset(data, "new_dataset",
data_storage_version="stable")
ds_legacy.data_storage_version
'2.0'
Or you can pass an explicit version:
ds_legacy = lance.write_dataset(data, "experimental_dataset",
data_storage_version="2.1")
ds_legacy.data_storage_version
'2.1'
Explicit versions will be a good choice if you know the exact version you want to use. Whereas stable is a good choice if you want to pick up the latest features, and next if you want to try out some unstable features. (We do not recommend next for any production use case. It's there for the purposes of collecting feedback on in-progress features.)
As we continue to develop the format and stabilize features, the stable and next version labels will move up versions. This will mean new tables created with these same tags will get new features as they are rolled out. However, existing tables will be kept at a fixed number version, resolved at the time you create the table. You will be able to migrate existing tables to new versions.
This new API was developed by LanceDB Engineer Weston Pace, who has been heading up our V2 file format development. (PR)
Distributed indexing
On the indexing front, we are exposing new APIs to scale up indexing performance. Our new distributed APIs allow breaking down the steps of creating an IVF_PQ ANN index and parallelizing some of them. By breaking down into steps, you can re-use certain intermediate results. This can be useful if you need to retry aborted jobs or restart execution with more resources.
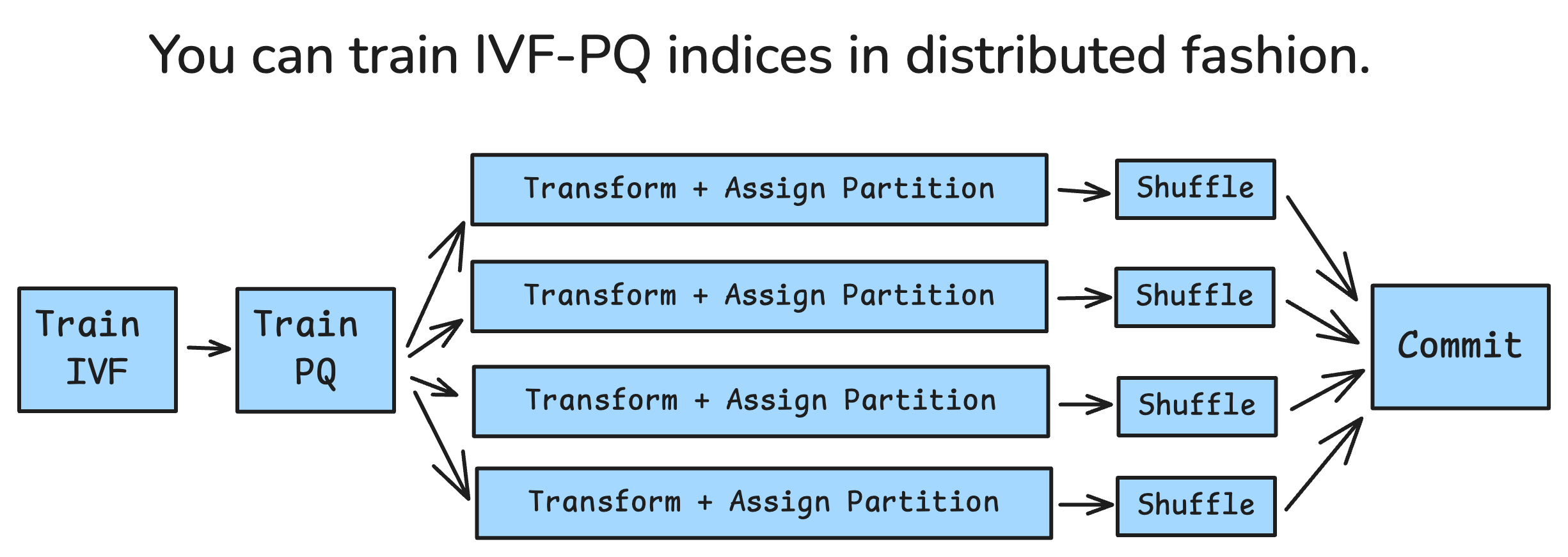
The most intensive steps are now able to be run in parallel. One of these is transforming the vectors from their lossless representation to their PQ-compressed representation. The other is assigning vectors to IVF partitions. These can be performed as a single transformation, and done completely in parallel. The other step that can be done in parallel is local shuffling vectors into partitions.
import pyarrow.compute as pc
dims = 128
nrows = 100_000
inner_vector = pc.cast(pc.random(nrows * dims), pa.float32())
vector = pa.FixedSizeListArray.from_arrays(inner_vector, dims)
table = pa.table({
"vector": vector,
"id": range(nrows),
})
# Using a small number of rows per file, to simulate a large dataset with
# many files.
ds = lance.write_dataset(table, 'test', mode='overwrite', max_rows_per_file=2_000)
Previously, creating an index was one step. This is still a good way to do indexing on a single machine, as it still uses multiple threads to take advantage of multi-core CPUs. Only when you want to do distributed indexing does the new API make sense.
# Indexing on single machine looks like:
ds.create_index(
column='vector',
index_type='IVF_PQ',
metric="cosine",
num_partitions=256,
num_sub_vectors=32,
)
The first two steps are creating the IVF and PQ models:
from lance.indices import IvfModel, PqModel, IndicesBuilder
builder = IndicesBuilder(ds, "vector")
ivf = builder.train_ivf(
num_partitions=256,
sample_rate=256,
distance_type="l2",
)
pq = builder.train_pq(ivf, num_subvectors=32)
Both of these can be saved to file. This is so they can be re-used, but also because in the distributed steps later they will need to be read by other processes.
import tempfile
import os
tmp_dir = tempfile.TemporaryDirectory()
ivf.save(os.path.join(tmp_dir.name, "ivf.index"))
pq.save(os.path.join(tmp_dir.name, "pq.index"))
Now we are at the step we can parallelize. As a simple example, we use a ThreadPoolExecutor, but this can be any sort of distributed process. (In fact, using a thread pool is a bad idea for real work. The transform_vectors step already has internal threading, so there's no point in trying to distribute multiple invocations across threads.) We use the fragments as the unit of parallelism, creating one task per fragment. Each of these becomes one output file.
from concurrent.futures import ThreadPoolExecutor
def transform_step(
ds_uri: str,
working_dir: str,
fragment_id: int
) -> str:
ds = lance.dataset(ds_uri)
fragment = ds.get_fragment(fragment_id)
ivf = IvfModel.load(os.path.join(working_dir, "ivf.index"))
pq = PqModel.load(os.path.join(working_dir, "pq.index"))
filename = "partition_" + str(fragment_id)
uri = os.path.join(working_dir, filename)
builder.transform_vectors(ivf, pq, uri, fragments=[fragment])
return filename
pool = ThreadPoolExecutor(max_workers=4)
fragment_ids = [frag.fragment_id for frag in ds.get_fragments()]
transformed_files = list(pool.map(
lambda fragment_id: transform_step(ds.uri, tmp_dir.name, fragment_id),
fragment_ids
))
Next, we can do the shuffle steps, which can also be done in parallel:
from typing import List
def shuffle_step(
working_dir: str,
transformed_files: List[str],
job_id: int,
):
ivf = IvfModel.load(os.path.join(working_dir, "ivf.index"))
return builder.shuffle_transformed_vectors(
transformed_files,
dir_path=tmp_dir.name,
ivf=ivf,
shuffle_output_root_filename=f"shuffled_{job_id}",
)
# Do one shuffle job per file
jobs = pool.map(
lambda x: shuffle_step(tmp_dir.name, [x[1]], x[0]),
zip(range(4), transformed_files)
)
shuffled_files = [file for job in jobs for file in job]The final step is to combine these shuffled files into an index file and commit that to the dataset:
builder.load_shuffled_vectors(
shuffled_files,
dir_path=tmp_dir.name,
ivf=ivf,
pq=pq
)
ds = ds.checkout_version(ds.latest_version)
ds.list_indices()[{'name': 'vector_idx',
'type': 'Vector',
'uuid': 'a657b964-d19e-41cc-ae8b-389cd2a95f9e',
'fields': ['vector'],
'version': 40,
'fragment_ids': {0,
1,
2,
3,
...
}}]
Given the complexity of the steps, this is a low-level API we don’t expect any but the most motivated power users to call. Instead, these power users may wrap them in distributed frameworks familiar to users, such as Ray and Spark. This will allow users to leverage their existing cluster infrastructure for their large-scale indexing jobs.
These features were developed by LanceDB engineer Weston Pace and our recent intern Raunak Shah.
Conclusion
These are a few of the major changes in v0.16.0 and v0.16.1. For a full list of changes, see our change logs:
- https://github.com/lancedb/lance/releases/tag/v0.16.0
- https://github.com/lancedb/lance/releases/tag/v0.16.1
In summary, the updates in Lance v0.16.1 focus on enhancing data management, improving performance, and facilitating distributed processing. The introduction of version tagging allows users to manage dataset versions more effectively. The ability to update subcolumns with merge_insert optimizes data updates, especially beneficial for large datasets. The stabilization of the V2 format ensures consistency and reliability in data storage. Lastly, the new distributed indexing APIs enable more efficient processing of large-scale datasets, paving the way for advanced and scalable data operations. These improvements not only streamline workflows but also expand the possibilities for users working with large and complex datasets.


